30 Nov 2017
The MYO-SEQ project: application of exome sequencing technologies to 1000 patients affected by limb-girdle weakness of unknown origin

Authors:
Katherine Johnson (katherine.johnson@ncl.ac.uk), Johnson, Katherine
John Walton Muscular Dystrophy Research Centre, MRC Centre for Neuromuscular Diseases, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
Töpf, Ana
John Walton Muscular Dystrophy Research Centre, MRC Centre for Neuromuscular Diseases, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
Bertoli, Marta
John Walton Muscular Dystrophy Research Centre, MRC Centre for Neuromuscular Diseases, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
Phillips, Lauren
John Walton Muscular Dystrophy Research Centre, MRC Centre for Neuromuscular Diseases, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
Blain, Alison
John Walton Muscular Dystrophy Research Centre, MRC Centre for Neuromuscular Diseases, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
Ensini, Monica
John Walton Muscular Dystrophy Research Centre, MRC Centre for Neuromuscular Diseases, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
Lek, Monkol
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA
Xu, Liwen
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA
Mullen, Thomas
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA
Valkanas, Elise
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA
MacArthur, Daniel G
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA
Straub, Volker
John Walton Muscular Dystrophy Research Centre, MRC Centre for Neuromuscular Diseases, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
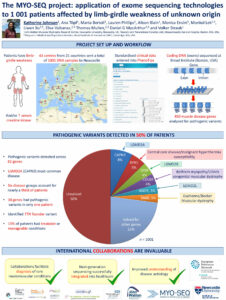
Muscular dystrophies are a heterogeneous group of rare genetic disorders that are characterised by progressive skeletal muscle wasting and weakness, and can directly precipitate premature mortality. Since up to 80% of these diseases have an underlying genetic cause, traditional clinical methodologies are often inadequate. Accordingly, we describe here the conclusion of the first phase of the MYO-SEQ project, an international research collaboration that applied targeted whole exome sequencing (WES) to the largest ever cohort of patients with undiagnosed proximal muscle weakness. We aimed to facilitate a clearer understanding of disease aetiology, enhance the diagnostic pathway and heighten the awareness of neuromuscular disorders – particularly limb-girdle muscular dystrophies (LGMDs). One thousand European participants presented with limb-girdle weakness and/or elevated creatine kinase activity. WES was performed using Illumina exome capture and a Picard-based processing pipeline. The variant call set was uploaded onto the Broad Institute of Harvard and MIT’s seqr platform and 450 muscle disease-associated genes were examined. Suspected pathogenic variants were identified in 50% of participants across 82 genes. Variants in CAPN3, RYR1, DYSF, COL6A1, 2 and 3, ANO5 and DMD accounted for over half of the solved patients. We also identified a TTN founder mutation in a Serbian sub-population. Genotype-phenotype correlations and inheritance patterns were expanded for many diseases. Crucially, we diagnosed over 140 patients with treatable or manageable conditions including hereditary cardiomyopathies, congenital myasthenic syndromes, metabolic disorders and ion channel diseases. We have facilitated the integration of next-generation sequencing technologies into healthcare. This study has significantly enhanced the capacity of standard clinical work-ups and has pioneered an accessible pathway to expedite future diagnoses.